Research Objectives:
This study explores the Intersection of Science and Technology in the digital era
Keywords:
Biology, Digital, Technology, Molecules, Bioinformatics’
Bio
Roxanne Boodhoo is an accomplished professional with a diverse and versatile background. Her extensive academic training has equipped her with a wide range of skills and knowledge, enabling her to excel in various roles. Roxanne is known for her strong work ethic, diligence, and commitment to undertaking any responsibilities assigned to her. She is deeply passionate about helping and supporting others, making her a compassionate and empathetic individual. Throughout her career, Roxanne has consistently demonstrated a dedication to making a positive impact, whether through her professional work or community involvement, striving to uplift those around her.
Abstract
In the digital era, technology has become a fundamental and cross-disciplinary component through which scientific knowledge is progressing and expanding exponentially. Biology in particular is undergoing a profound transformation thanks to the development of bioinformatics and computational methods. Bioinformatics has changed the study of biology and has made it possible to store, process, analyse and extract useful information from large amounts of data, such as those obtained with gene sequencing. The support of the digital component is increasingly essential for experimental biology, from molecules to ecosystems. The world of today is becoming increasingly complex due to the integration and interaction between disciplines, such as computer science and biology. Bioinformatics is an innovative discipline that inherits traditional biological approaches while incorporating biological data and computational resources. Bioinformatics is located at the centre of modern highthroughput experimentation. One of the most prominent tools in bioinformatics is the BLAST algorithm, used to search for sequences in databases. Living organisms are the result of a continuous evolutionary process and are characterised by the inheritance of genetic information and population dynamics. One of the main challenges of bioinformatics is how to characterise, classify and understand both the biological and the specific organisational features that underlie the molecular function of a nucleic acid or protein molecule. Bioinformatics delves into annotative, structural, functional, evolutionary and regulatory genomics.
Introduction
Biology is a technique-centred field, but it is increasingly emphasising moving towards problems (Gunaga et al., 2020). This is particularly important in biomedicine were using the old focus on techniques rather than overall investigation can lead to over-diagnosis, and treatments that are highly effective for some patients and not at all for others. Overcoming these challenges efficiently will require more efficient multi-disciplinary, multi-method substrates of interoperation that include multiple different data sharing and collaborative scientific solutions. On all of these issues, libraries are the key to developing plans associated with our shared resources. And digital libraries are where the transformations that JBiD is proposing must have their eventual, symphonic confluence.
The Journal of Biomedical Discovery and Collaboration offers a venue for a wide 1. Introduction array of interdisciplinary discoveries, methods, and techniques in order to support these three points and more (R Smalheiser, 2006). The completion of the human genome project marked the formal connection of science to digital technology. Ever since, biomedical research has been transitioning to being a fully information science (M. Thampi, 2009). Yet, the problems of biology do not need computer scientists, or laboratory and clinical investigators alone. They also need data scientists, social scientists, and ethicists, and most importantly, scientific and clinical leadership capable of integrating these different investigation methods into a singular approach. The formal requirement for interdisciplinarity in 21st century biomedicine is that problems are more structured than they are in basic science. And it is now widely recognised that the leadership skillset for problem solving is different from that required (and largely rewarded) in the prior century of basic science approach to hypothesis testing and support.
This research study takes the cursive movement of the transformation of the biosciences in this “digital era” as its starting point (Lee & Helgesson, 2022). Our concern is the rich landscape of digital transformations under way within contemporary biology; transformations bound up with new and old valuation cultures; experimentation and forms-of-life; shifts in time, uncertainty and automation; co and post-modelled worlds; and, throughout, biological matter caught in the entangled semiotics of infinity and the ongoing work of becoming-fixed. The figure of the pathway is used here as an allegory for the transformation of contemporary biology – from codes to data to matter to machines – in order to surface the multiple technoscientific heritage of the present and to insist on the need for similarly multi-perspectival sociology of biology & technology if the moving contours of contemporary biology are to be adequately captured.
Technological convergence in the biosciences is prompting new questions about longstanding sociological concerns surrounding digitization in scientific practice. Where many familiar narratives have focused on digital tools as an empty vessel for human knowledge, a renewed and updated sociology of the digital in biology might better account for the situated and coordinated nature of digitalization within this field (A. Peters et al., 2021). Rather than focusing on the “entrance” of technology into human practice, here we more informatively seek to study the ongoing cointegration of human and machine practices. Thinking about biology and biotechnology in this way, as part of the same systemic transformation, may enable us to better understand how digital work is multiple, multifaceted and, indeed, always already sociotechnical.
Aim
Naturally, in soil, a chain of host-symbiont crosses talks to each other and find the best matchups. Research in rhizobia was spearheaded by Jostein Goksoyr and Kornelius Lindstrom. Metabolic and environmental persistence attributes pave the way for the fitness and genetics analysis. Throughout the world several such gene sequences are stored, and RDP is one such database that provides the tool as well as their respective avenues of these gene sequences pertaining to 16S rRNA, atpD, recA, dnaK, glnA, and rpoB of different isolates as characterised. A repository of 10 such Bhagwant University rhizobial gene sequences has been deposited into DDBJ/EMBL/GenBank by our research group with detailed classification and a proper evolutionary relationship drawn. It has been constructed on the basis of either of two measures such as: the homologous > e-10,67 bp alignment length by aligning the kmers set or the ITER registry of primary protein structure patterns.
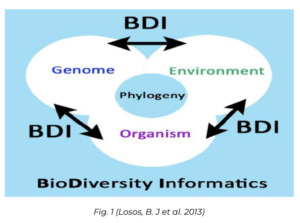
Rhizobia, a group of soil bacteria of great agricultural significance, associate with leguminous plants and contribute to improved crop production and ecosystem health because of the process of root nodule formation in leguminous plants, such as beans, peas, soybeans, and peanuts (B. Losos et al., 2013). So, these leguminous plants are able to convert atmospheric nitrogen into ammonia. This process is called nitrogen fixing symbiosis and the microorganism is known as Rhizobium, which primarily modulates herbaceous legumes. Currently, worldwide there are about 600 million ha of agricultural land restored in BNF of leguminous species having the potential for cultivation. It has been reported that India has approximately more than 8.5 million ha under pulse cultivation out of which more than 1.3 million ha has been estimated to have the possibility for legume cultivation. Fig 1 highlights that evolutionary biology is undergoing a transformation due to the growing availability of extensive data on genomic variation, organisms, and environmental factors.
Bioinformatic technologies increasingly facilitate research in taxonomy, systematics, and phylogenetics. In this project, we aim to apply bioinformatics in taxonomy, systematics, and phylogenetics of selected Rhizobia species (M. Thampi, 2009). Bioinformatics approaches such as sequence alignment, molecular phylogeny, and in silico DNA-DNA hybridization (DDH) capabilities will be used to accurately delineate Rhizobia species. Wholegenome sequences of sixteen Rhizobium strains will be explored for systematics, and phylogenetic and relative DDH analyses. This project should aid in the identification of novel indigenous Rhizobial species suitable for the repatriation, exchange of the economy, and conservation of all bioresources of Union Country. It is a foregone fact that these Rhizobial taxonomic activities are time-consuming and share high risk due to increases in the number of newly isolated additional indigenous Rhizobium species from Union Country which demand laborious biochemical profiling.
Method
Rapid advance in technology has accelerated development of biology from an empirical science to a data-centric one. The intersection of computing and biology has been recently showing an entirely new potential connecting many applications of computer science and biotechnology. Biological computing has been an exciting area, with the practical language of DNA substrates leading the grounding development (Akula & Cusick, 2009). To use DNA to implement computation has stimulated people’s thinking about DNA computing and related biological computing. Over time, many new disciplines, such as programming with genetic bits, DNA walkers and the emerging biocomputing field, have been inspired by the particular computational model, making the software and hardware different from earlier biological computing. DNA computing advantages were related to DNA perfect memory, miniaturization, concurrent computing, negligible thermal noise and low cost-operation. It is notable that using DNA as a processor and memory can be negligible anyway at an atomic level for miniaturization or cause a degradation, thereby these physical processes are instruction and output of a biocomputer conversely. Also, since no error repairing occurs in instruction reading from DNA, a negligible thermal noise and data processing by stochastic diffusion may even make users believe to have used traditional computing’s incalculable random sequence considerations.
In recent years, biology has transformed from an empirical discipline to natural sciences, especially thanks to advanced technologies such as highthroughput data collection tools (B. Losos et al., 2013). Among these tools, highthroughput sequencing has revolutionised many sub-fields of biology. The large-scale and low-cost DNA sequencing performed by machines can produce gigabytes of genomic data within minutes, replacing traditional Sanger sequencing of DNA. This sea change in high-throughput sequencing technology has led to the sequencing of numerous viral and cellular genomes, transforming the biological science from an empirical and study of small samples to a data-driven one, in which an observation in an experiment can often be presented and interpreted in terms of big data, such as cellular RNA-Seq and treatment with chemical probes or genetic siRNA or CRISPR libraries. In the last decade, machine learning techniques were applied on the big genomic data to predict gene composition, illness risks and drug efficacy, among many other examples (Gunaga et al., 2020).
Results and Discussion
As we expected from the beginning of our discussion, the battery and the solar panel, which we use to replace the classical boiler, guarantee a higher computational power when the boiler needs to spend more than 156 hours of work. Furthermore, from the economic point of view, the battery and the solar panel are more convenient when a computational step has to be spent more than 322 hours. We merge these considerations by plotting an hourly cost versus the total number of required computational steps. The cost difference between the three system versions is expected to decrease when the total number of the computational steps grows, as the boiler becomes less and less competitive. To emphasise our considerations, we have found a pass between the boiler and the battery, in the plane of the two quantities. All the considered theoretical, technological and economic aspects confirm our previous conclusions (Patra et al., 2022). In summary, we have shown a way to experimentally measure the energy gap between the initial and final state, using digital biochemistry in an ideal scenario. This way, we have derived a figure of merit for each system, making it possible to rank them. They could easily compare physical energy with information energy, measuring systems’ potential impact in terms of environmental respect and technological innovation. For example, a list of environments could be offered to the final users, one for each system, next to the output progressional. Also, this benchmark could be employed within the framework of a computational multi-objective optimisation, where instead the thermodynamic optimisation is ended, the selection evolves, as a final system, according to the best Pareto-optimal set. Following these thoughts, we recall on a waiting list all the file status and actions to be performed outside the time lapse of 3 days with the prices in terms of the two quantities.
In this section, we focus only on the theoretical experiment and analyse the resources used. To obtain the energy consumption and the costs, we consider the boiler’s, battery, and solar panel costs, the electricity price, the technoeconomic and environmental analysis of the solar panel, and the ideal cases of the initial state of the system.
Conclusion
The domain of biology is entering an era of big data, where multi-omic and systems-scale research are becoming industry standard and open scientific culture is no longer a potential exception (E. Thessen & J. Patterson, 2011) (K. Rennstich, 2018). As a result, big databases, large datasets, and software tools are becoming a natural part of scientific life. In this article, we focused on the following topics relevant to the current state of the data-intensive landscape of life sciences. We discussed the reasons for the emergence of big data in biology as well as the ways different biological disciplines cope with the changes. Finally, we presented a list of focal points relevant to making big data in biology work at a level that will be effective and useful. Especially advocated were: (i) thoughtful design of databases and interfaces, in a way that would address cognitive, physical, and positively affect scientific community experience; (ii) breaking away from the idea of universal solutions, at the same time advocating solutions directed toward a specific domain or laboratory practice; (iii) reform of data integration operations from the ground up, on which we still depend to build theories and collect and annotate data; (iv) building open-science oriented software tools from the current most popular modelling technologies, in a way that would favour open data standards, software transparency, and universal compatibility. Finally, we quickly reviewed more general problems like the profound changes needed in education in biology, and more generally in the training of interdisciplinary skills. We raised the problems and perspectives of personal responsibility and openness in laboratory-to-laboratory data management to avoid the overfitting era of the scientific discourse. We also mentioned diversity in teamwork, gap analysis in multi-omic data publications, and the rapid change of the perception of network biology behind. And all those in an interdisciplinary way. In the era of big data, it is essential that biological knowledge is widely disseminated and effectively managed (Zitnik et al., 2023). During the last two decades, the field has seen the emergence of new ways of thinking and new methods for network-based analysis. As has been the case in other fields, more and more elaborate discussions, reflections and discussions on the next steps that network biology must take can be observed, and our network can significantly move us away from the understanding of animal models and the interpretation of hypothesisbased experimental outputs. These “blessings,” however, can be misused leading to loss of credibility and major financial waste. If we learn to predict, aren’t we creating a world of self-prophetic pseudo-autistic machine learning models? As described by Yu, prediction has now outreached explanation and this is reflected across disciplines and scales. Effective use of network models and integrative systems-level analysis brings several technically significant challenges as well as skills for an interdisciplinary success story. In this conclusion, we briefly review current state of the field, the directions of which we think network science in life sciences is starting to move, and where the social diversity in computational biology can help.
References
A. Peters, M., Jandrić, P., & Hayes, S. (2021). Biodigital Philosophy, Technological Convergence, and Postdigital Knowledge Ecologies. ncbi. nlm.nih.gov
Akula, B. & Cusick, J. (2009). Biological Computing Fundamentals and Futures. [PDF]
E. Thessen, A. & J. Patterson, D. (2011). Data issues in the life sciences. ncbi.nlm.nih.gov
Gunaga, S., Vinod Prabhu, V., M. S., R., Kulkarni, A., & C. Iyer, N. (2020). Selection of Robust Digital Communication Techniques for the Vehicle to Vehicle Communication. [PDF]
K. Rennstich, J. (2018). The world system in the information age: structure, processes, and technologies. osf.io
Lee, F. & Helgesson, C. F. (2022). Styles of Valuation: References Algorithms and Agency in High- throughput Bioscience. osf.io
Losos, B. J., J. Arnold, S., Bejerano, G., D. Brodie, E., Hibbett, D., E. Hoekstra, H., P. Mindell, D., Monteiro, A., Moritz, C., Allen Orr, H., A. Petrov, D., S. Renner, S., E. Ricklefs, R., S. Soltis, P., & L. Turner, T. (2013). Evolutionary Biology for the 21st Century. ncbi.nlm.nih.gov
M. Thampi, S. (2009). Introduction to Bioinformatics. [PDF]
Patra, S., Mukherjee, A., Mukherjee, A., S. Vidhyadhiraja, N., Taraphder, A., & Lal, S. (2022). Frustration shapes multichannel Kondo physics: a star graph perspective. [PDF]
R Smalheiser, N. (2006). Launching the Journal of Biomedical Discovery and Collaboration. ncbi.nlm.nih. gov
Zitnik, M., M. Li, M., Wells, A., Glass, K., Morselli Gysi, D., Krishnan, A., M. Murali, T., Radivojac, P., Roy, S., Baudot, A., Bozdag, S., Z. Chen, D., Cowen, L., Devkota, K., Gitter, A., Gosline, S., Gu, P., H. Guzzi, P., Huang, H., Jiang, M., Nesibe Kesimoglu, Z., Koyuturk, M., Ma, J., R. Pico, A., Pržulj, N., M. Przytycka, T., J. Raphael, B.,Ritz, A., Sharan, R., Shen, Y., Singh, M., K. Slonim, D., Tong, H., Holly Yang, X., Yoon, B. J., Yu, H., & Milenković, T. (2023). Current and future directions in network biology. [PDF]


